Saccharomyces cerevisiae Pathway/Genome Database |
 |
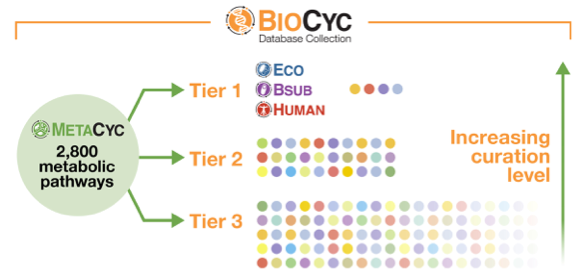
YeastCyc is a Pathway/Genome Database of the model eukaryote Saccharomyces cerevisiae S288c. In addition to genomic information, the database contains metabolic pathway, reaction, enzyme, and compound information, which has been manually curated from the scientific literature. The curation process includes periodic updates to the metabolites, reactions and pathways from the current version of MetaCyc to ensure compatibility with other members of the MetaCyc family of databases. YeastCyc has been curated from 47,000 publications. For details on recent curation, please click here.
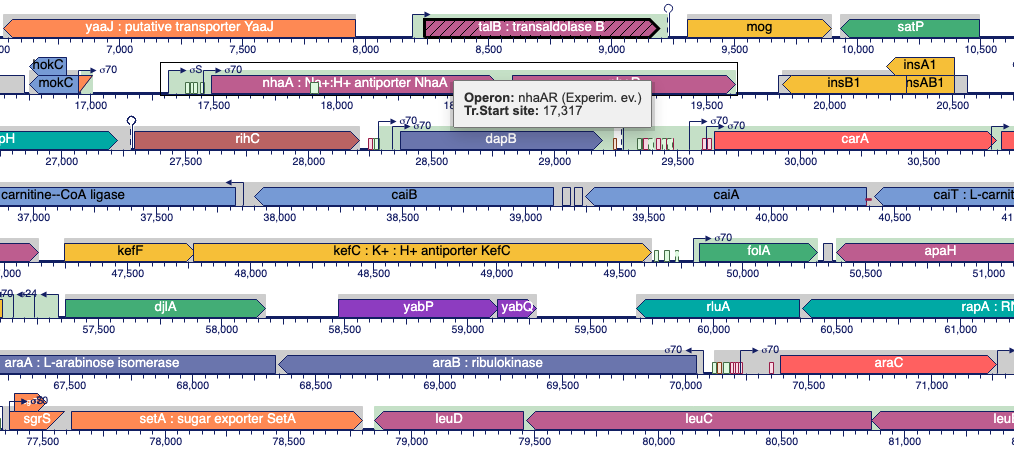
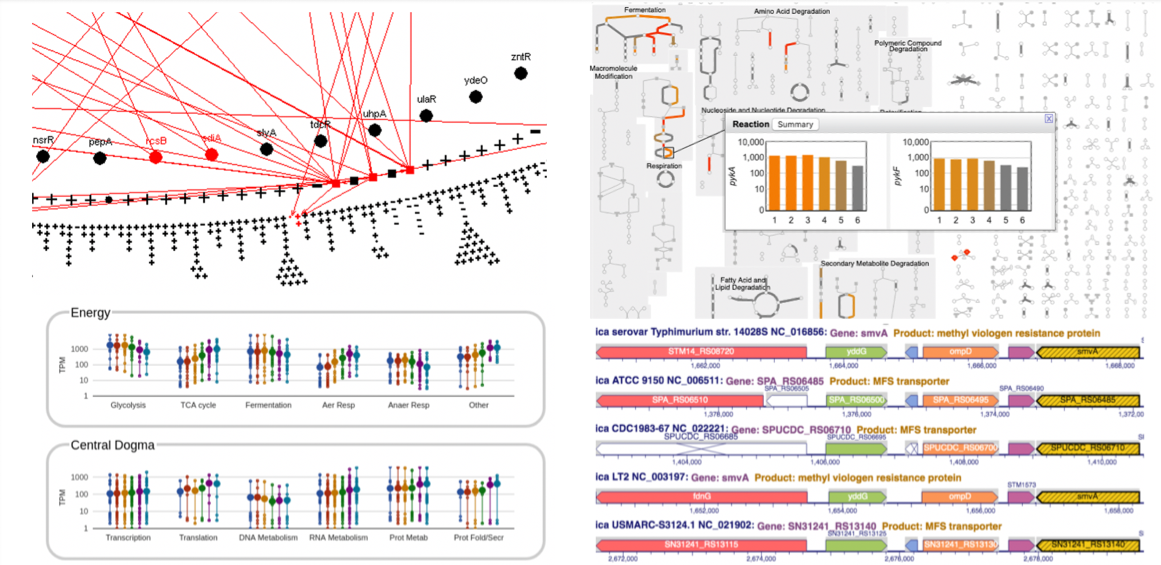
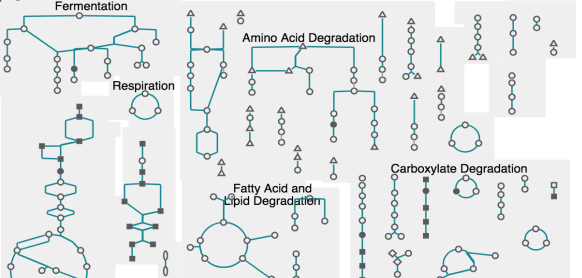
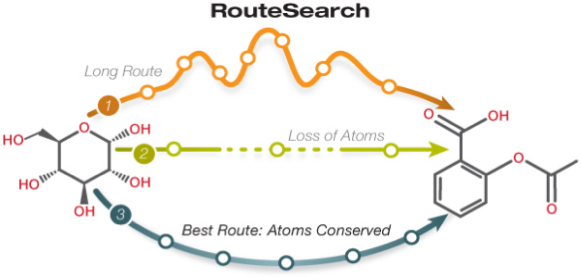
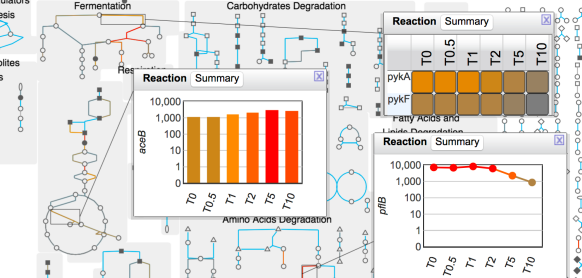
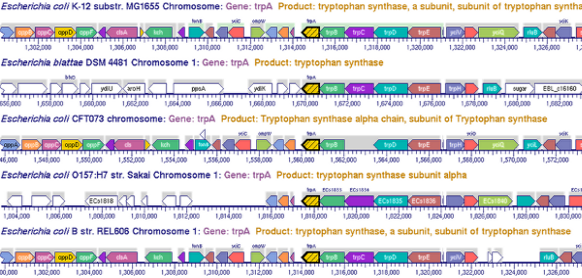
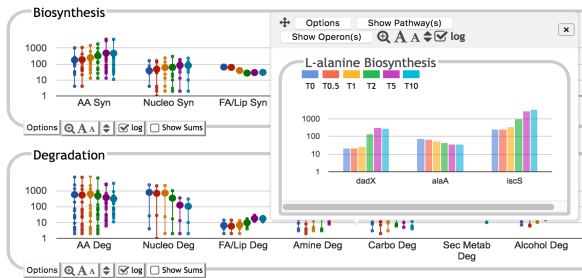
This site includes extensive retrieval, visualization and analysis tools, including a genome browser and a metabolic map diagram. There are tools for analysis of gene expression, metabolomics and ChIP-chip data. You can search and align sequences for Saccharomyces cerevisiae and other microbial genomes and perform comparative analysis. You can store groups of genes and pathways into a SmartTable, and then browse, analyze and share with other users.
What people are saying
See more BioCyc testimonials
"BsubCyc is a tool of the utmost value."

Paul Babitzke
Prof. of Biochemistry
& Molecular Biology
"My lab uses these resources on a daily basis."

Patricia Kiley,
Professor and Chair,
Dep't. of Biomolecular Chemistry
"We rely on BioCyc's Gene Pages and Overview Diagrams almost daily."

Arkady Khodursky
Assoc. Prof. Biochemistry
"We use BioCyc and MetaCyc extensively to investigate the metabolic and regulatory processes of organisms we study."

William Cannon, Team Lead
Computational Biology
"BioCyc is the go-to resource of knowledge and tools for Ginkgo scientists."

"BioCyc is a tremendous resource for pathway analysis in metabolomics."

Art Edison, Dept of Genetics
"We make extensive use of the BioCyc full metabolic network diagram for omics data analysis."

Timothy J. Donohue, Director
"I have not found another database that has a better interface than BioCyc."

Gary B. Huffnagle, Professor
Microbiology and Immunology
Learning Library
Tutorial Videos
Tutorial #1: Introduction to BioCyc
Tutorial #2: Introduction to SmartTables
Tutorial #3: Zoomable Metabolic Map, Comparative Tools, Regulatory Network
Tutorial #4: Omics Data Analysis
Tutorial #5: Pathway Collages
Tutorial #6: Creating a Pathway/Genome Database
- Part 1A: Introduction to Database Building and Pathologic (14:04)
- Part 1B: Building a Database: Detailed Pathologic Example (23:53)
- Part 2A: General Editing Strategies (8:00)
- Part 2B: Creating and Editing Reactions and Compounds (17:32)
- Part 2C: Updating Proteins, Citations, GO Terms, and Enzymatic Reactions (26:10)
- Part 2D: Making and Editing Pathways (9:42)